
Overview
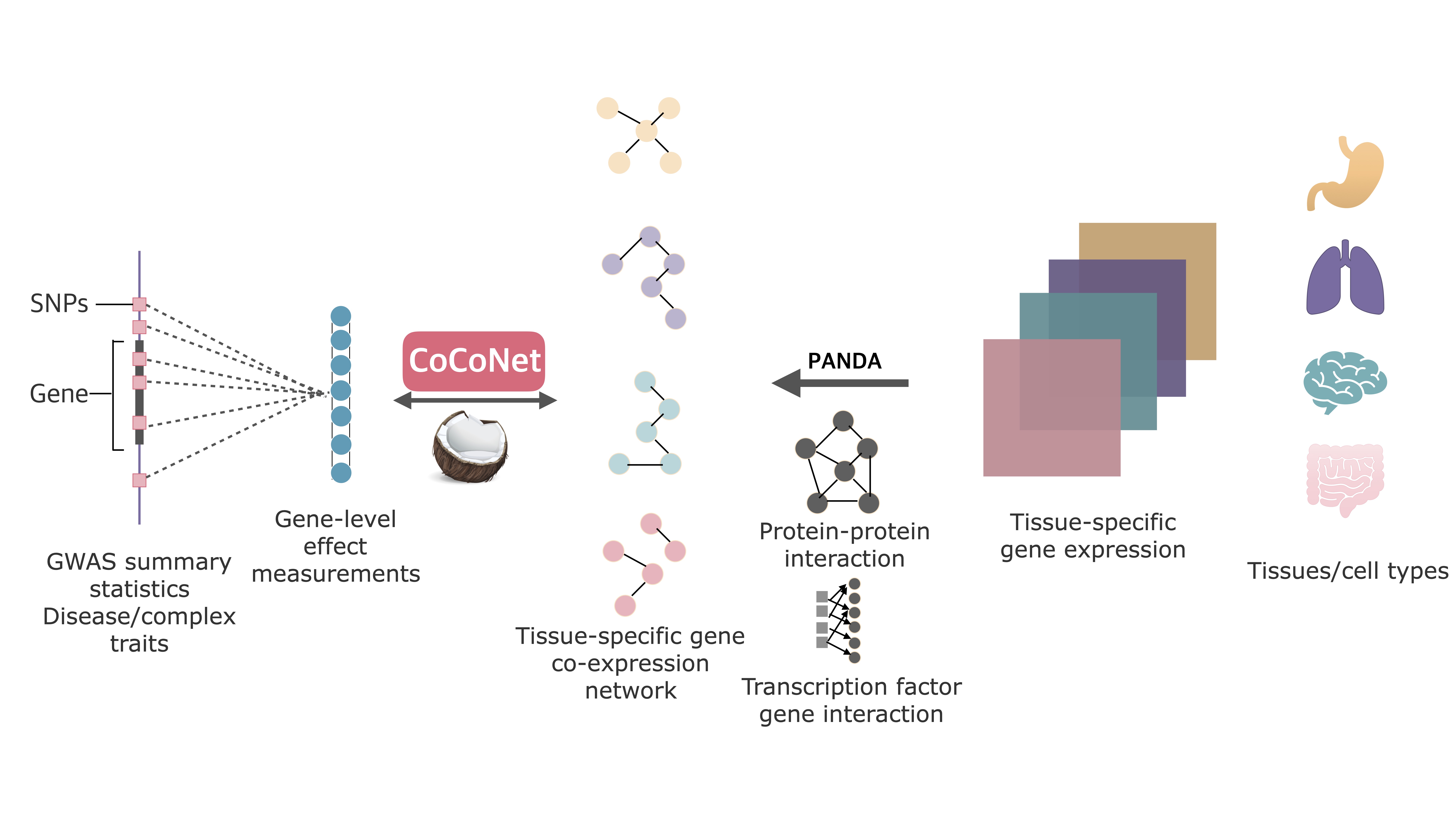
CoCoNet is an efficient method to facilitate the identification of trait-relevant tissues or cell types. We apply CoCoNet for an in-depth analysis of four neurological disorders and four autoimmune diseases, where we integrate the corresponding GWASs with bulk RNAseq data from 38 tissues and single cell RNAseq data from 10 cell types.
CoCoNet incorporates tissue-specific gene co-expression networks constructed from either bulk or single cell RNA sequencing (RNAseq) studies with GWAS data for trait-tissue inference. In particular, CoCoNet relies on a covariance regression network model to express gene-level effect sizes for the given GWAS trait as a function of the tissue-specific co-expression adjacency matrix. With a composite likelihood-based inference algorithm, CoCoNet is scalable to tens of thousands of genes.
Paper:
Leveraging Gene Co-expression Patterns to Infer Trait-Relevant Tissues in Genome-wide Association Studies
https://doi.org/10.1371/journal.pgen.1008734
Tutorial of CoCoNet
Package Website: CoCoNet Tutorial