Abstract
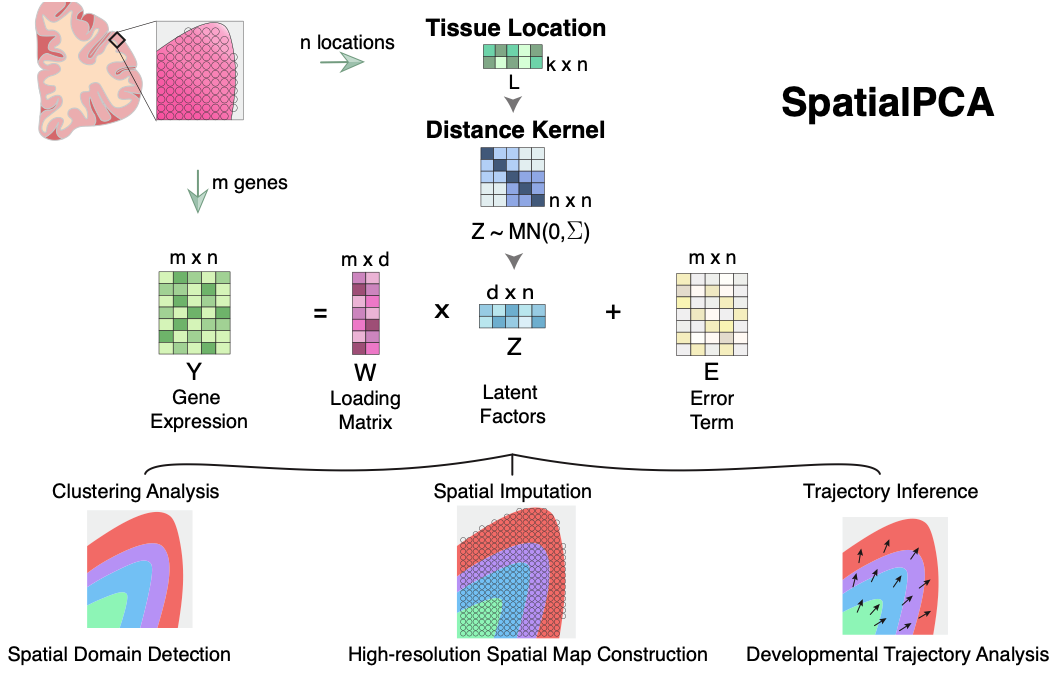
Overview SpatialPCA is a spatially aware dimension reduction method that aims to infer a low dimensional representation of the gene expression data in spatial transcriptomics.
Materials and Methods SpatialPCA builds upon the probabilistic version of PCA, incorporates localization information as additional input, and uses a kernel matrix to explicitly model the spatial correlation structure across tissue locations.
Results In the real data applications, SpatialPCA identifies key molecular and immunological signatures in a newly detected tumor surrounding microenvironment, including a tertiary lymphoid structure that shapes the gradual transcriptomic transition during tumorigenesis and metastasis. In addition, SpatialPCA detects the past neuronal developmental history that underlies the current transcriptomic landscape across tissue locations in the cortex.
Conclusion SpatialPCA is computationally efficient, easily scalable to spatial transcriptomics with tens of thousands of spatial locations and thousands of genes. We have illustrated the benefits of SpatialPCA for spatial transcriptomics visualization, spatial domain detection, trajectory inference on the tissue, as well as high-resolution spatial map construction.
Paper:
https://www.nature.com/articles/s41467-022-34879-1
Tutorial of SpatialPCA
SpatialPCA Tutorial
