SpatialPCA
Lulu Shang, Xiang Zhou
2022-01-27
Last updated: 2022-08-13
Checks: 2 0
Knit directory: Tutorial/
This reproducible R Markdown analysis was created with workflowr (version 1.7.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version d8f1e8e. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: analysis/_main.html
Untracked files:
Untracked: code/Simulation.R
Untracked: code/SpatialPCA.R
Untracked: code/SpatialPCA_EstimateLoading.R
Untracked: code/SpatialPCA_SpatialPCs.R
Untracked: code/SpatialPCA_buildKernel.R
Untracked: code/SpatialPCA_highresolution.R
Untracked: code/SpatialPCA_utilties.R
Untracked: data/LIBD_sample9.RData
Untracked: data/Puck_200115_08_count_location.RData
Untracked: data/ST_data.RData
Untracked: data/Tumor_data.RData
Untracked: data/Vizgen_Merfish_count_location.RData
Untracked: data/slideseq.rds
Unstaged changes:
Modified: analysis/ST.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were
made to the R Markdown (analysis/index.Rmd) and HTML
(docs/index.html) files. If you’ve configured a remote Git
repository (see ?wflow_git_remote), click on the hyperlinks
in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| html | 7964c20 | shangll123 | 2022-03-22 | Build site. |
| Rmd | 251c36f | shangll123 | 2022-03-22 | Publish SpatialPCA tutorial |
| Rmd | 9a0d220 | shangll123 | 2022-01-06 | wflow_git_commit(all = TRUE) |
| Rmd | 4a0f267 | shangll123 | 2022-01-06 | Start workflowr project. |
Welcome to SpatialPCA!
Overview
SpatialPCA is a spatially aware dimension reduction method that aims to infer a low dimensional representation of the gene expression data in spatial transcriptomics. SpatialPCA builds upon the probabilistic version of PCA, incorporates localization information as additional input, and uses a kernel matrix to explicitly model the spatial correlation structure across tissue locations.
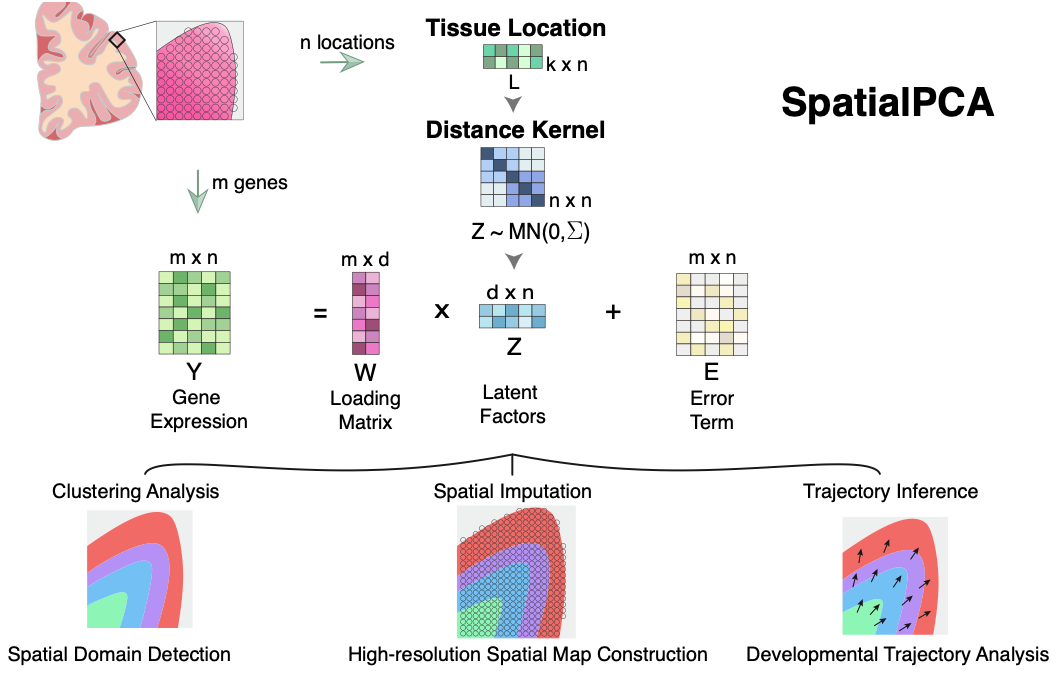
Please read our paper at Nature Communications:
Spatially Aware Dimension Reduction for Spatial Transcriptomics