Abstract
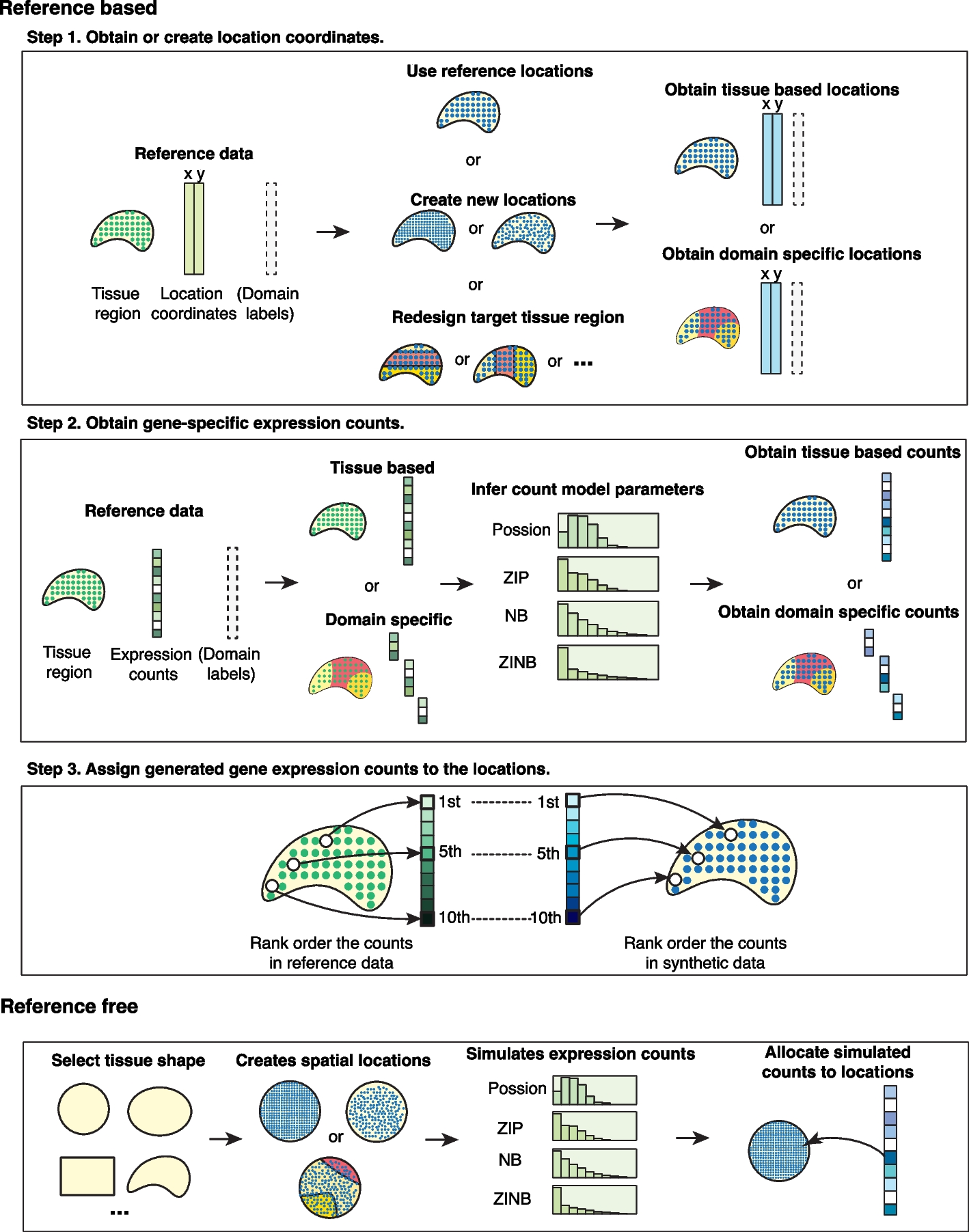
Overview Spatially resolved transcriptomics (SRT)-specific computational methods are often developed, tested, validated, and evaluated in silico using simulated data. Unfortunately, existing simulated SRT data are often poorly documented, hard to reproduce, or unrealistic. Single-cell simulators are not directly applicable for SRT simulation as they cannot incorporate spatial information.
Materials and Methods SRTsim, an SRT-specific simulator for scalable, reproducible, and realistic SRT simulations. SRTsim not only maintains various expression characteristics of SRT data but also preserves spatial patterns.
Results We illustrate the benefits of SRTsim in benchmarking methods for spatial clustering, spatial expression pattern detection, and cell-cell communication identification.
Conclusion In conclusion, SRTsim is an effective simulation tool for SRT studies. SRTsim provides scalable, reproducible, and realistic simulations, while preserving expression characteristics and spatial patterns. We demonstrated the utility of SRTsim in evaluating the performance of spatial clustering methods, spatial expression analysis methods, and cell-cell communication identification methods.
Paper:
https://genomebiology.biomedcentral.com/articles/10.1186/s13059-023-02879-z
Tutorial of SRTsim
SRTsim Tutorial