Abstract
Background Proteomics, the large-scale analysis of proteins, is contributing greatly to understanding gene function in the postgenomic era. However, disease protein ranking using shotgun proteomics data has not been fully evaluated. In this study, we prioritized disease-related proteins by integrating the protein-protein interaction (PPI) network and protein differential expression profiles from colon and rectal cancer (CRC) or breast cancer (BC) proteomics.
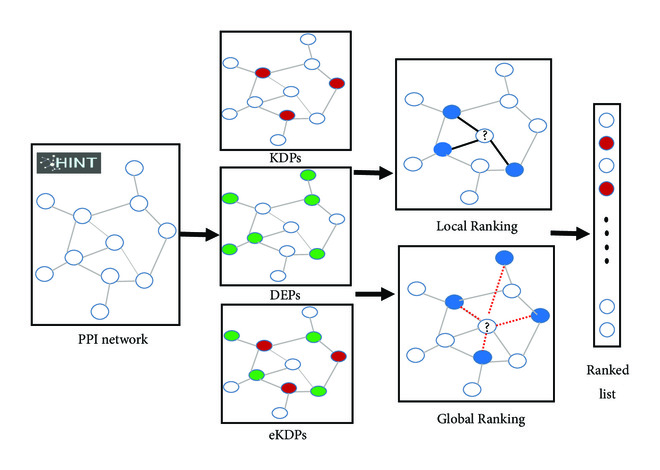
Materials and Methods We applied Local Ranking (LR) and Global Ranking (GR) methods in network with three kinds of protein sets as a priori knowledge, which were known disease proteins (KDPs) that were collected from the Online Mendelian Inheritance in Man (OMIM) database, differentially expressed proteins (DEPs), and the collection of KDPs and their direct neighborhood with differential expression (eKDPs).
Results The cross-validations showed that GR method outperformed LR method while using eKDPs as the initial training showed significantly higher accuracy compared to using the other two a priori sets. And then we validated the top ranked proteins using RNAi-based loss-of-function screens in the DepMap database. The results showed that 75% of top 20 proteins in CRC are necessary for tumor survival.
Conclusion
In summary, the network-based Global Ranking with protein differential expression can efficiently prioritize cancer-related proteins and discover new candidate cancer genes or proteins.